Note
Click here to download the full example code
04. Plot contrast¶
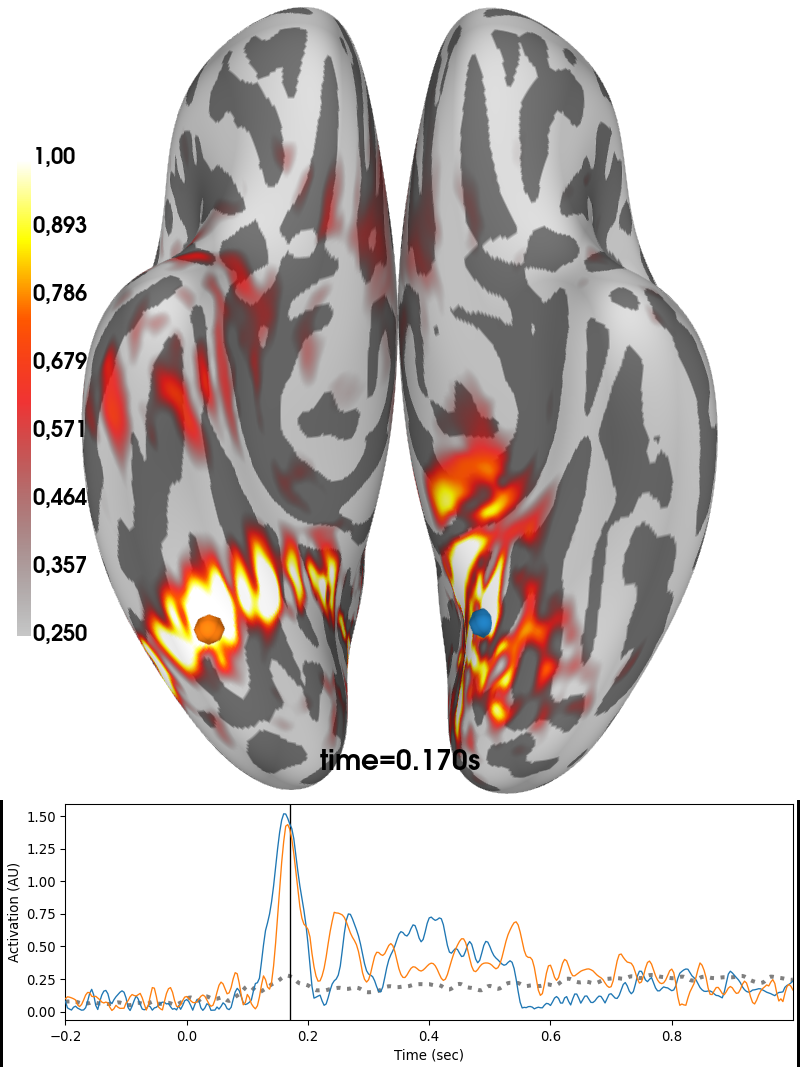
Group average of dSPM solutions obtained by 03. Compute inverse solution for the contrast between both types of faces together and scrambled at 170 ms poststimulus. The image was produced by subtracting normalized solutions of faces to the ones of scrambled.
# Authors: Annalisa Pascarella <a.pascarella@iac.cnr.it>
# License: BSD (3-clause)
# sphinx_gallery_thumbnail_number = 1
import os
import json
import pprint
import os.path as op
import numpy as np
import mne
# Read experiment params as json
params = json.load(open("params.json"))
pprint.pprint({'parameters': params})
data_type = params["general"]["data_type"]
subject_ids = params["general"]["subject_ids"]
NJOBS = params["general"]["NJOBS"]
session_ids = params["general"]["session_ids"]
new_name_condition = params["inverse"]["new_name_condition"]
is_short = params["general"]["short"] # # to analyze a segment of data
if "data_path" in params["general"].keys():
data_path = params["general"]["data_path"]
else:
data_path = op.expanduser("~")
print("data_path : %s" % data_path)
subjects_dir = op.join(data_path, params["general"]["subjects_dir"])
src_estimation_wf_name = 'source_dsamp_short_reconstruction_dSPM_aparc' \
if is_short else 'source_dsamp_full_reconstruction_dSPM_aparc'
morph_stc_path = \
op.join(data_path, src_estimation_wf_name,
'_subject_id_{sbj}', 'morph_stc')
os.environ['QT_API'] = 'pyqt5'
fig_path = op.join(data_path, 'figures')
if not os.path.isdir(fig_path):
os.mkdir(fig_path)
# PLot
stc_condition = list()
for cond in new_name_condition:
stcs = list()
for subject in subject_ids:
out_path = morph_stc_path.format(sbj=subject)
stc = mne.read_source_estimate(
op.join(out_path, 'mne_dSPM_inverse_morph-%s' % (cond)))
stcs.append(stc)
data = np.average([np.abs(s.data) for s in stcs], axis=0)
stc = mne.SourceEstimate(data, stcs[0].vertices,
stcs[0].tmin, stcs[0].tstep, 'fsaverage')
del stcs
stc_condition.append(stc)
data = stc_condition[0].data / np.max(stc_condition[0].data) + \
stc_condition[2].data / np.max(stc_condition[2].data) - \
stc_condition[1].data / np.max(stc_condition[1].data)
data = np.abs(data)
stc_contrast = mne.SourceEstimate(
data, stc_condition[0].vertices, stc_condition[0].tmin,
stc_condition[0].tstep, 'fsaverage')
# stc_contrast.save(op.join(fig_path, 'stc_dspm_difference_norm'))
lims = (0.25, 0.75, 1)
clim = dict(kind='value', lims=lims)
brain_dspm = stc_contrast.plot(
views=['ven'], hemi='both', subject='fsaverage', subjects_dir=subjects_dir,
initial_time=0.17, time_unit='s', background='w',
clim=clim, foreground='k', backend='auto')
brain_dspm.save_image(op.join(fig_path, 'dspm-contrast.png'))
'''
brain_dspm = stc_contrast.plot(
views='ven', hemi='lh', subject='fsaverage', subjects_dir=subjects_dir,
initial_time=0.17, time_unit='s', background='w',
clim=clim, foreground='k', backend='matplotlib')
'''
{'parameters': {'general': {'NJOBS': 1,
'data_path': '/home/pasca/Science/workshop/PracticalMEEG/ds000117/derivatives/meg_derivatives',
'data_type': 'fif',
'session_ids': ['01', '02'],
'short': False,
'subject_ids': ['sub-01'],
'subjects_dir': '/home/pasca/Science/workshop/PracticalMEEG/ds000117/FSF'},
'inverse': {'condition': ['face/famous',
'scrambled',
'face/unfamiliar'],
'events_id': {'face/famous/first': 5,
'face/famous/immediate': 6,
'face/famous/long': 7,
'face/unfamiliar/first': 13,
'face/unfamiliar/immediate': 14,
'face/unfamiliar/long': 15,
'scrambled/first': 17,
'scrambled/immediate': 18,
'scrambled/long': 19},
'method': 'dSPM',
'new_name_condition': ['famous',
'scrambled',
'unfamiliar'],
'parcellation': 'aparc',
'snr': 3.0,
'spacing': 'oct-6',
'tmax': 1.0,
'tmin': -0.2,
'trans_fname': '*-trans.fif'},
'preprocessing': {'ECG_ch_name': 'EEG063',
'EoG_ch_name': ['EEG061', 'EEG062'],
'down_sfreq': 300,
'h_freq': 40,
'l_freq': 0.1,
'reject': {'grad': 4e-10, 'mag': 4e-12},
'variance': 0.999}}}
data_path : /home/pasca/Science/workshop/PracticalMEEG/ds000117/derivatives/meg_derivatives
Using pyvistaqt 3d backend.
"\nbrain_dspm = stc_contrast.plot(\n views='ven', hemi='lh', subject='fsaverage', subjects_dir=subjects_dir,\n initial_time=0.17, time_unit='s', background='w',\n clim=clim, foreground='k', backend='matplotlib')\n"
Total running time of the script: ( 0 minutes 2.866 seconds)