Note
Click here to download the full example code
02. Compute ERP¶
This workflow mainly call the
ephypype pipeline
computing N170 component from cleaned EEG data. The first Node of the workflow
(Extract events Node Node) extracts the events from raw data. The events
are saved in the Node directory.
In the ERP Node the raw data are epoched accordingly to events
extracted in Extract events Node.
The evoked datasets are created by averaging the different conditions specified
in json file.
# Authors: Annalisa Pascarella <a.pascarella@iac.cnr.it>
# License: BSD (3-clause)
# sphinx_gallery_thumbnail_number = 2
Import modules¶
The first step is to import the modules we need in the script. We import mainly from nipype and ephypype packages.
import os
import os.path as op
import json
import pprint # noqa
import ephypype
import nipype.pipeline.engine as pe
from nipype.interfaces.utility import Function
from ephypype.nodes import create_iterator, create_datagrabber
from ephypype.pipelines.preproc_meeg import create_pipeline_evoked
from ephypype.datasets import fetch_erpcore_dataset
Let us fetch the data first. It is around 90 MB download.
directory /home/pasca/workshop already exists
Define data and variables¶
Let us specify the variables that are specific for the data analysis (the
main directories where the data are stored, the list of subjects and
sessions, …) and the variable specific for the particular pipeline
(events_id, baseline, …) in a
json file
(if it is does work, try to go on the github page, and right-click “Save As” on the Raw button)
# Read experiment params as json
params = json.load(open("params.json"))
pprint.pprint({'parameters': params["general"]})
data_type = params["general"]["data_type"]
subject_ids = params["general"]["subject_ids"]
NJOBS = params["general"]["NJOBS"]
session_ids = params["general"]["session_ids"]
# data_path = params["general"]["data_path"]
# ERP params
ERP_str = 'ERP'
pprint.pprint({'ERP': params[ERP_str]})
events_id = params[ERP_str]['events_id']
condition = params[ERP_str]['condition']
baseline = tuple(params[ERP_str]['baseline'])
events_file = params[ERP_str]['events_file']
t_min = params[ERP_str]['tmin']
t_max = params[ERP_str]['tmax']
{'parameters': {'NJOBS': 1,
'data_path': '',
'data_type': 'eeg',
'session_ids': ['N170'],
'subject_ids': ['016']}}
{'ERP': {'baseline': [None, 0],
'condition': ['faces', 'car'],
'events_file': '*_filt_ica-eve.fif',
'events_id': {'car': 2, 'faces': 1},
'tmax': 0.8,
'tmin': -0.2}}
Extract events¶
The first Node of the workflow extract events from the raw data. The events are extracted using the function events_from_annotations of MNE-python on raw data. The events are saved in the Node directory.
def get_events(raw_ica, subject):
'''
First, we get the ica file from the preprocessing workflow directory, i.e.
the cleaned raw data. The events are extracted from raw annotation and are
saved in the Node directory.
'''
print(subject, raw_ica)
import mne
rename_events = {
'201': 'response/correct',
'202': 'response/error'
}
for i in range(1, 180 + 1):
orig_name = f'{i}'
if 1 <= i <= 40:
new_name = 'stimulus/face/normal'
elif 41 <= i <= 80:
new_name = 'stimulus/car/normal'
elif 101 <= i <= 140:
new_name = 'stimulus/face/scrambled'
elif 141 <= i <= 180:
new_name = 'stimulus/car/scrambled'
else:
continue
rename_events[orig_name] = new_name
raw = mne.io.read_raw_fif(raw_ica, preload=True)
events_from_annot, event_dict = mne.events_from_annotations(raw)
faces = list()
car = list()
for key in event_dict.keys():
if rename_events[key] == 'stimulus/car/normal':
car.append(event_dict[key])
elif rename_events[key] == 'stimulus/face/normal':
faces.append(event_dict[key])
merged_events = mne.merge_events(events_from_annot, faces, 1)
merged_events = mne.merge_events(merged_events, car, 2)
event_file = raw_ica.replace('.fif', '-eve.fif')
mne.write_events(event_file, merged_events)
return event_file
Specify Nodes¶
Before to create a workflow we have to create the nodes that define the workflow itself. In this example the main Nodes are
infosourceis a Node that just distributes valuesdatasourceis a DataGrabber Node that allows the user to define flexible search patterns which can be parameterized by user defined inputsextract_eventsis a Node containing the function Extract eventsERP_pipelineis a Node containing the pipeline created bycreate_pipeline_evoked()
Infosource and Datasource¶
We create a node to pass input filenames to
infosource = create_iterator(['subject_id', 'session_id'],
[subject_ids, session_ids])
# ``datasource`` node to grab data. The ``template_args`` in this node iterate
# upon the values in the infosource node
ica_dir = op.join(
data_path, 'preprocessing_workflow', 'preproc_eeg_pipeline')
template_path = "_session_id_%s_subject_id_%s/ica/sub-%s_ses-%s_*filt_ica.fif"
template_args = [['session_id', 'subject_id', 'subject_id', 'session_id']]
datasource = create_datagrabber(ica_dir, template_path, template_args)
Extract events Node¶
Then, we define the Node that encapsulates get_events function
extract_events = pe.Node(
Function(input_names=['raw_ica', 'subject'],
output_names=['event_file'],
function=get_events),
name='extract_events')
ERP Node¶
Finally, we create the ephypype pipeline computing evoked data which can be connected to these nodes we created.
Specify Workflows and Connect Nodes¶
Now, we create our workflow and specify the base_dir which tells
nipype the directory in which to store the outputs.
ERP_pipeline_name = ERP_str + '_workflow'
main_workflow = pe.Workflow(name=ERP_pipeline_name)
main_workflow.base_dir = data_path
We then connect the output of infosource node to the one of
datasource. So, these two nodes taken together can grab data.
main_workflow.connect(infosource, 'subject_id', datasource, 'subject_id')
main_workflow.connect(infosource, 'session_id', datasource, 'session_id')
We connect the output of infosource and datasource to the input
of extract_events node
main_workflow.connect(datasource, 'raw_file', extract_events, 'raw_ica')
main_workflow.connect(infosource, 'subject_id', extract_events, 'subject')
Finally, we connect the output of infosource, datasource and
extract_events nodes to the input of ERP_pipeline node.
main_workflow.connect(infosource, 'subject_id',
ERP_workflow, 'inputnode.sbj_id')
main_workflow.connect(datasource, 'raw_file',
ERP_workflow, 'inputnode.raw')
main_workflow.connect(extract_events, 'event_file',
ERP_workflow, 'inputnode.events_file')
Run workflow¶
After we have specified all the nodes and connections of the workflow, the
last step is to run it by calling the run method. It’s also possible to
generate static graph representing nodes and connections between them by
calling write_graph method.
main_workflow.write_graph(graph2use='colored') # optional
'/home/pasca/workshop/ERP_CORE/ERP_workflow/graph.png'
Take a moment to pause and notice how the connections here correspond to how we connected the nodes.
import matplotlib.pyplot as plt # noqa
img = plt.imread(op.join(data_path, ERP_pipeline_name, 'graph.png'))
plt.figure(figsize=(6, 6))
plt.imshow(img)
plt.axis('off')
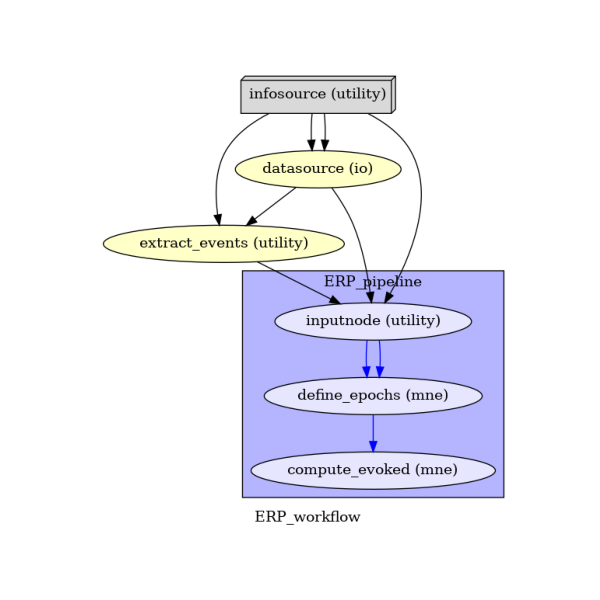
(-0.5, 535.5, 594.5, -0.5)
Finally, we are now ready to execute our workflow.
main_workflow.config['execution'] = {'remove_unnecessary_outputs': 'false'}
# Run workflow locally on 1 CPU
main_workflow.run(plugin='LegacyMultiProc', plugin_args={'n_procs': NJOBS})
<networkx.classes.digraph.DiGraph object at 0x7fdef9f2e410>
Plot results¶
import mne # noqa
import matplotlib.pyplot as plt # noqa
from ephypype.gather import get_results # noqa
evoked_files, _ = get_results(main_workflow.base_dir,
main_workflow.name, pipeline='compute_evoked')
for evoked_file in evoked_files:
print(f'*** {evoked_file} ***\n')
ave = mne.read_evokeds(evoked_file)
faces, car = ave[0], ave[1]
gfp_faces = faces.data.std(axis=0, ddof=0)
gfp_car = car.data.std(axis=0, ddof=0)
# compare conditions
contrast = mne.combine_evoked([faces, car], weights=[1, -1])
gfp_contrast = contrast.data.std(axis=0, ddof=0)
# Reproducing the MNE-Python plot style seen above
fig, ax = plt.subplots()
ax.plot(faces.times, gfp_faces * 1e6, color='blue')
ax.plot(car.times, gfp_car * 1e6, color='orange')
ax.plot(contrast.times, gfp_contrast * 1e6, color='green')
ax.legend(['Faces', 'Car', 'Contrast'])
fig.show()
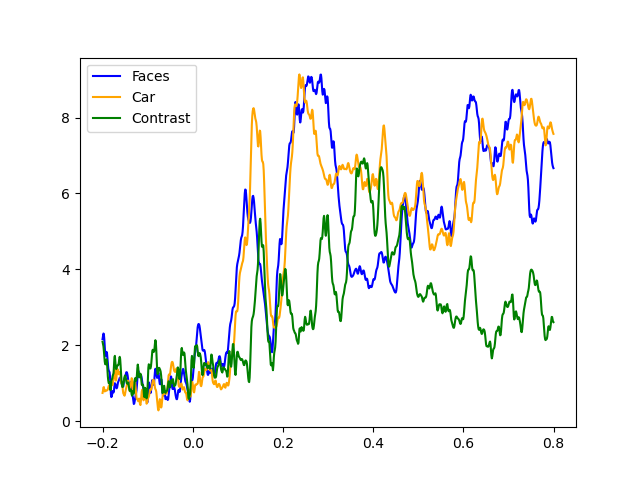
*** /home/pasca/workshop/ERP_CORE/ERP_workflow/ERP_pipeline/_session_id_N170_subject_id_016/compute_evoked/sub-016_ses-N170_task-N170_eeg_filt_ica-ave.fif ***
Reading /home/pasca/workshop/ERP_CORE/ERP_workflow/ERP_pipeline/_session_id_N170_subject_id_016/compute_evoked/sub-016_ses-N170_task-N170_eeg_filt_ica-ave.fif ...
Read a total of 1 projection items:
Average EEG reference (1 x 29) active
Found the data of interest:
t = -200.20 ... 799.80 ms (faces)
0 CTF compensation matrices available
nave = 70 - aspect type = 100
Projections have already been applied. Setting proj attribute to True.
Loaded Evoked data is baseline-corrected (baseline: [-0.200195, 0] sec)
Read a total of 1 projection items:
Average EEG reference (1 x 29) active
Found the data of interest:
t = -200.20 ... 799.80 ms (car)
0 CTF compensation matrices available
nave = 76 - aspect type = 100
Projections have already been applied. Setting proj attribute to True.
Loaded Evoked data is baseline-corrected (baseline: [-0.200195, 0] sec)
Total running time of the script: ( 0 minutes 8.560 seconds)