Neuropycon¶
Neuropycon is an open-source multi-modal brain data analysis kit which provides Python-based pipelines for advanced multi-thread processing of fMRI, MEG and EEG data, with a focus on connectivity and graph analyses. Neuropycon is based on Nipype, a tool developed in fMRI field, which facilitates data analyses by wrapping many commonly-used neuro-imaging software into a common python framework.
Neuropycon project includes two different packages:
- ephypype based on MNE python includes pipelines for electrophysiology analysis
- graphpype based on radatools includes pipelines for graph theoretical analysis of neuroimaging data
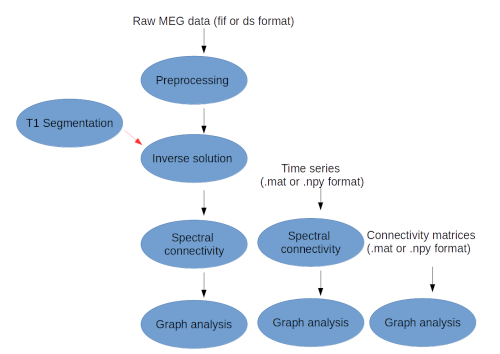
Neuropycon provides a very common and fast framework to develop workflows for advanced analyses, in particular defines a set of different pipelines that can be used stand-alone or as lego of a bigger workflow: the input of a pipeline will be the output of another pipeline.
For each possible workflow the input data can be specified in three different ways:
- raw MEG data in .fif and .ds format
- time series of connectivity matrices in .mat (Matlab) or .npy (Numpy) format
- connectivity matrices in .mat (Matlab) or .npy (Numpy) format
Each pipeline based on nipype engine is defined by nodes connected together, where each node maybe wrapping of existing software (as MNE-python modules or radatools functions) as well as providing easy ways to implement function defined by the user.
graphpype¶
Neuropycon project for graph analysis, can be used from ephypype and nipype.
The graphpype package provides the following pipelines:
- the conmat_to_graph pipeline runs the graph computation and graph-theoretical tools over connectivity matrices.
- the inv_ts_to_graph pipeline runs the spectral connectivity and the graph computation over time series.
- the nii_to_graph pipeline provide a script to compute connectivity matrices and graphs computations from prepocessed functional MRI.
- the inv_ts_to_bct_graph pipeline provide example scripts to compute graph metrics (so far, KCore) using bctpy (Brain Connectivity Toolbox).
Installation¶
graphpype works with python3
$ pip install https://api.github.com/repos/neuropycon/graphpype/zipball/master
Or with pip:
$ pip install graphpype
Radatools¶
You should add all the directories from radatools to the PATH env variable:
- Download radatools sotware:
http://deim.urv.cat/~sergio.gomez/radatools.php#download
- Download and extract the zip file
- Add following lines in your .bashrc:
For radatools 3.2¶
RADA_PATH=/home/david/Tools/Software/radatools-3.2-linux32
(replace /home/david/Tools/Software by your path to radatools)
export PATH=$PATH:$RADA_PATH/01-Prepare_Network/
export PATH=$PATH:$RADA_PATH/02-Find_Communities/
export PATH=$PATH:$RADA_PATH/03-Reformat_Results
export PATH=$PATH:$RADA_PATH/04-Other_Tools/
For radatools 4.0¶
RADA_PATH=/home/david/Tools/Software/radatools-4.0-linux64
(replace /home/david/Tools/Software by your path to radatools)
export PATH=$PATH:$RADA_PATH/Network_Tools
export PATH=$PATH:$RADA_PATH/Network_Properties
export PATH=$PATH:$RADA_PATH/Communities_Detection
export PATH=$PATH:$RADA_PATH/Communities_Tools
For radatools 5.0¶
RADA_PATH=/home/david/Tools/Software/radatools-5.0-linux64
(replace /home/david/Tools/Software by your path to radatools)
export PATH=$PATH:$RADA_PATH/Network_Tools
export PATH=$PATH:$RADA_PATH/Network_Properties
export PATH=$PATH:$RADA_PATH/Communities_Detection
export PATH=$PATH:$RADA_PATH/Communities_Tools